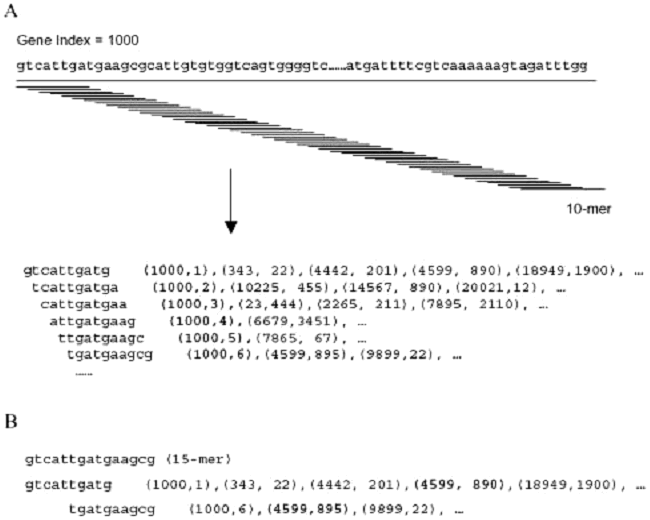
Oligonucleotide Probe SelectionDespite the widespread use of oligonucleotide-based microarrays, the research community has not yet implemented a set of standardized criteria by which oligonucleotide probes are designed and used. While important measures have been taken to standardize the format in which microarray data is reported, standardization of probe design would further facilitate microarray data comparison. Because the development of oligonucleotide probes has been driven by the commercial sector, only a few reported methods of probe design and selection have been made publicly available. In an effort to provide an open-source algorithm for oligonucleotide selection, the MGH Microarray Core has chosen to use an oligonucleotide selection algorithm developed by Xiaowei Wang and Brian Seed from the MGH Bioinformatics group of Parabiosys. The algorithm, termed OligoPicker, applies a variety of simple filters to candidate sequences derived from the open reading frame. Based upon the NCBI protein database Genpept, corresponding DNA coding sequences are retrieved based on the coding sequence (CDS) feature in the Genpept file. Redundant coding sequences are clustered giving higher priority to RefSeq sequences (excluding RefSeq genes predicted from genomic contig assembly process), orthologous sequences and longer coding sequences. Each DNA coding sequence was aligned against all other sequences in the dataset using BLAST. Sequences with at least 96% identity to the query sequence and with an alignment equal to or greater than the query length were considered redundant and removed. Additionally, probe sequences must minimally meet a defined Tm criteria of 74°C. Based upon this criteria, a starting pool of 60,000 murine sequences was simplified to 20,030 gene clusters. Because cross-hybridization will occur between regions with contiguous or perfectly matched bases, the primary filter rejects potential probes that contain ≥ 15 perfect base-matches with any other input sequence as demonstrated in Figure 1.
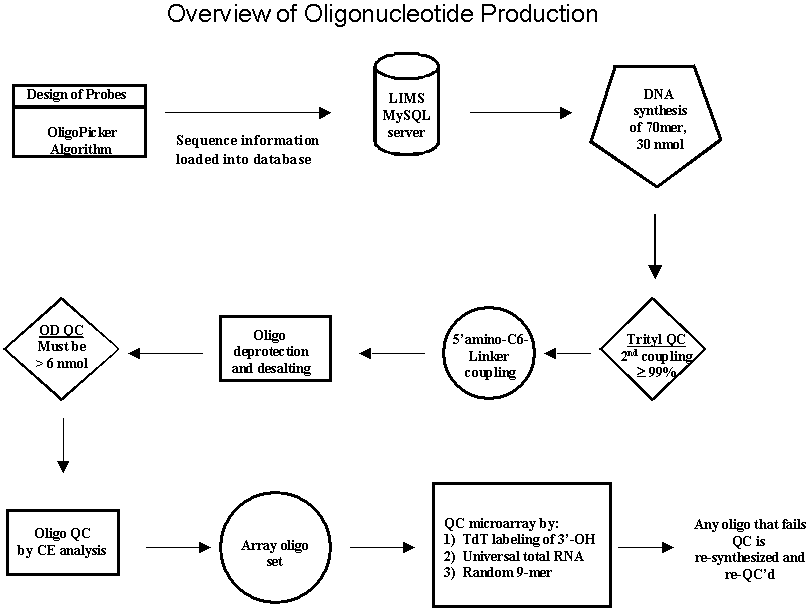
After application of the 15-base contiguous match filter, candidate probe sequences are screened further by applying a series of low-complexity, RNA, self-annealing, and BLAST score filters. If a sequence passes all of these filters, a selected probe sequence should uniquely represent its corresponding input sequence whilst minimizing the possibility of cross-hybridization. In certain instances, when a family of proteins is highly homologous at the mRNA level, a single probe sequence is chosen that uniquely represents that protein family. With this publicly available algorithm in-hand, the MGH Microarray Core takes advantage of the high-throughput DNA synthesis capabilities of the MGH Oligonucleotide Core to provide custom designed microarrays, domain specific microarrays, and customized high-density genomic microarrays. To date we have designed genomic probe sets for the mouse, human, rat, cow, chicken and pig, and completed synthesis of a 14,609 mouse genomic probe set. We are currently synthesizing an upgrade to the mouse set for complete genome coverage and will begin to produce the human probe set. The mouse set is available for purchase at cost to those academic institutions searching for a low-cost alternative to industrial oligonucleotide probe sets. Oligonucleotide Synthesis and Quality ControlSince its inception, the MGH Microarray Facility strongly advocated that a microarray core should have the ability to synthesize oligonucleotide probes in-house. Not only does this flexibility permit the Microarray Facility to design and fabricate custom microarrays, but also allows the Facility to support investigators who work on potential model systems for which no commercial oligonucleotide probe sets exist. In achieving this goal, the MGH Oligonucleotide Core has established a high-throughput synthesis program capable of producing 288 5'-amino modified 70mer probes per day on both a Polyplex synthesizer (Genomic Solutions) and a Mermade V synthesizer (BioAutomation) .At this level of synthetic capacity, 30,000 probes, one representing every putative gene in the human genome, could be synthesized in less than five months. A goal of the Microarray Facility is the ability to in-house manufacture oligonucleotide probes for the production of custom microarrays. In order to coordinate and monitor these large synthetic projects, the MGH Oligonucleotide Core, in collaboration with software engineers in the Bioinformatics group, has designed and implemented the necessary database infrastructure to support high-throughputsynthetic efforts. This laboratory information management system (LIMS) assists in oligonucleotide ordering, oligonucleotide annotation, tracking, and oligonucleotide quality control. Quality control is of paramount importance to the Oligonucleotide Core. The amino-modified 70mer probes used for microarray fabrication are exhaustively analyzed to ensure probes are full-length and in good yield by the following quality control metric (Figure 2). Oligonucleotide syntheses are monitored by the trityl release following the first coupling and the last coupling. The step-wise repetitive yield is computed based on the total number of couplings. Oligonucleotides with less than a 99% coupling efficiency are rejected. After the final detritylation, the 70mer probe is 5'-modified with a C6 amino-linker using standard phosphoramidite coupling chemistry. The oligonucleotides are subsequently deprotected, desalted, and the optical density of each amino-modified 70mer measured at 260 nm.
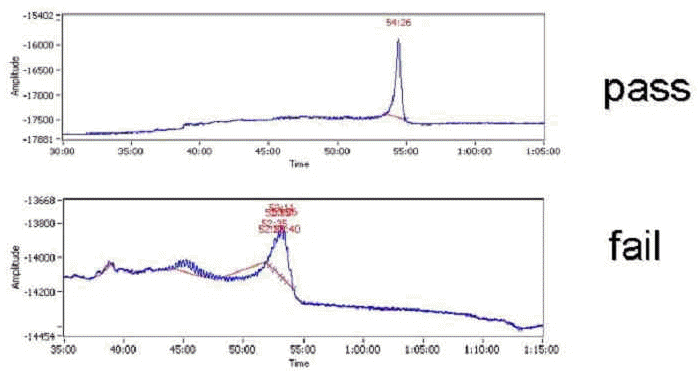
To monitor heterogeneity in oligonucleotide length, each oligonucleotide is subjected to capillary electrophoresis (Figure 3) using a 96-well plate compatible instrument (CombiSep). After analysis, capillary electrophoresis data from each individual amino-modified 70mer probe is uploaded into the LIMS. The LIMS makes several quality control calls based upon the full-length peak retention time and the area associated with that peak. The first QC call is made based upon the overall yield of the probe. Using Beer's law, the LIMS calculates the molar amount of full-length product based upon the fraction of full-length product in the sample and the optical density. If any sample fails to have at least 6 nmol of the amino-modified probe, the oligonucleotide is rejected. Additionally, no other peak in the chromatogram may be 20% of the total peak area. After quality control assessment, the modified 70mer probes that have successfully passed QC are collected. The collection process is performed by the staff of the MGH Automation Core using a Beckman/Sagian system, a sophisticated liquid handling robot with the ability to "cherry-pick" the QC'd probes and pool them into 384-well plates.
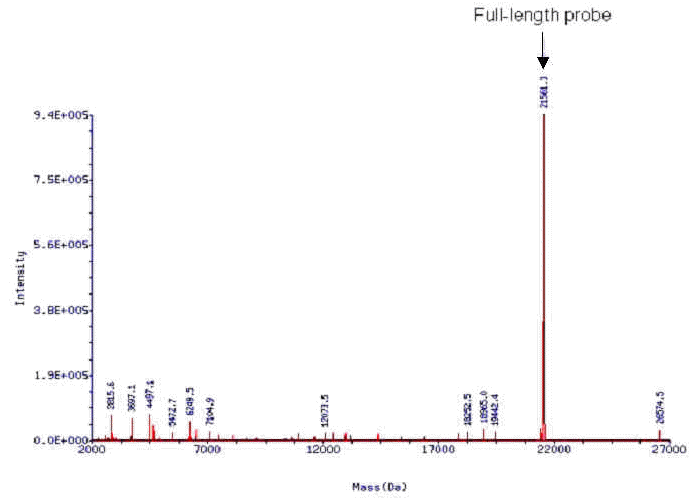
In an effort to further improve oligo QC methods, we have been collaborating with Novatia to develop methods for high resolution oligonucleotide analysis. Novatia has developed a high throughput electrospray ionization mass spectrometry technique called Oligo HTCS. The Oligo HTCS system is the first LC/MS system to automate oligonucleotide mass spectral analysis. Oligonucleotide samples are first desalted during initial LC, and then fragmented upon electrospray ionization. The fragmentation pattern generated for each oligonucleotide sample is then deconvoluted using Novatia's ProMass software allowing the reconstitution of the full-length oligonucleotide and its relative abundance in the sample.The instrumentation has the ability to support a throughput of up to10 x 96-well plates perday and enables the Oligonucleotide Core to immediately assess the quality of recently synthesized oligos so that oligonucleotide production is not interrupted. More importantly, the Oligo HTCS allows the characterization of long (70-120mers) oligos that are typically used in the manufacture of DNA microarrays, something that is nearly impossible using standard MALDI-ToF. Many companies that sell microarray oligo sets have purposefully limited the length of their oligonucleotide probes to ≥50 bases so that MALDI-ToF could be performed. We have sought alternative methods to this approach to limit decreases in probe sensitivity with decreasing probe length. We have found experimentally that the Oligo HTCS can rapidly and effectively QC 70mers (Figure 4) that are used on our DNA microarrays and are plannng for this method to be our future oligo QC protocol.
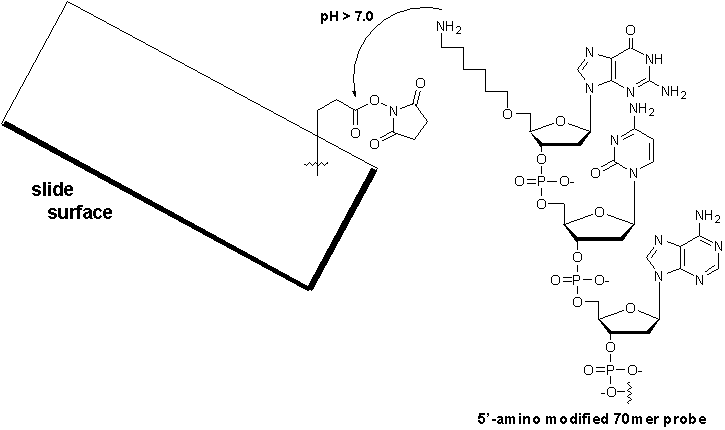
Microarray substrate and oligonucleotide couplingMicroarrays are typically printed on a glass surface to allow visualization of the hybridized, labeled targets. Reviewing the literature, glass slides have continued to be the preferred support for immobilizing probes for reasons including availability, transparency, low fluorescence, and resistance to high temperature. While poly-L-lysine coated slides are still popular substrates due to their low cost and generally good results, we have found higher levels of background associated with these slides in comparison to commercially available substrates. The Microarray Core has compared several slide supports and found that activated CodeLink slides (Amersham) gave a markedly higher signal-to-noise ratio improving the overall sensitivity by 16% when compared to poly-L-lysine coated slides and aldehyde-modified glass slides. CodeLink slides contain a gel-like polymer surface that is derivatized with an N-hydroxysuccinimide ester. The N-hydroxysuccinimide ester is the reactive moiety that mediates the specific coupling to the 5'-amine on the modified probe and results in the formation of a chemically stable amide bond (Figure 5). This coupling chemistry is advantageous. First, covalent bond-formation links the probes in an unidirectional manner. Second, because the probes contain a 6-carbon linker at the 5'-terminus, the probes are directed away from the microarray surface allowing greater access to the labeled target during hybridization.
Microarray Fabrication and Quality ControlUsing our initial probe set, we have successfully developed a standardized set of protocols for both microarray fabrication and microarray quality control that keeps costs minimal, reduces lot-to-lot variability and improves microarray performance. Presently, investigators at Massachusetts General Hospital, Brigham and Women's Hospital and Harvard Medical School are using these novel microarrays that have been printed, post-processed, and quality assessed by our facility. Once a probe set has been synthesized and arrayed into 384-well format, microarray production begins with the deposition of the 5'-amino modified probes onto the surface of activated Codelink slides (Amersham). In preparation for spotting, the oligonucleotide print sets are diluted to 20 µM in water, and 5.0 µL of each probe is transferred robotically into arrayer-friendly V-bottom 384-well polystyrene plates (Genetix) and dried. Immediately prior to printing, the probes are dissolved in 150 mM sodium phosphate buffer, pH 8.5 to allow the base catalyzed coupling of the 5'-amino modified probe to the slide surface (Figure 5). Probe spotting is performed using an Omnigrid Arrayer (GeneMachines), an xyz-axis robot capable of printing 100 microarrays per print run. Following spotting, the slides are incubated in 75% relative humidity for 14-16 h to hydrate the salt crystals and allow the probes to couple to the slide surface. After probe immobilization, the arrays are post-processed to deactivate any remaining NHS-esters using a blocking solution containing the primary amine, ethanolamine. The arrays are thoroughly washed and rinsed, and then dessicated until use. To check the overall print quality after a print run, the MGH Microarray Facility has developed a simple yet effective method of evaluating spot morphology, spot uniformity, print artifacts, and post-processing artifacts. After post-rocessing, the first and last slide of each print run are sacrificed for quality control purposes. Using terminal deoxynucleotidyl transferase (TdT), the 3'-OH moiety of each probe is labeled with Cy3-dCTP. To facilitate mixing during incubation, the microarrays are enzymatically labeled on a GeneTAC hybridization station (Genomic solutions). Briefly, 24 units of TdT, 2 µM of Cy3-dCTP are mixed in 1x TdT reaction buffer (124 µL total volume). The labeling reaction is added to the microarray and incubated at 37 °C for 2 h with agitation. After labeling, the microarray is washed with 2xSSC containing 0.1% SDS, followed by 2x SSC, and finally water. The MGH Microarray Facility uses a combination of TdT labeling, random 9mer hybridization, and universal total RNA hybridization for assessing array performance. If a probe fails to generate a minimum signal upon TdT labeling, the probe is resynthesized. If a probe is detected by the TdT assay, yet fails both the random 9mer and universal total RNA hybridizations, the oligo is redesigned, resynthesized, and reassayed. These quality control measures help ensure that both array quality and performance remain high.
With NHLBI-PGA support, we have been able to generate methods of producing in-house manufactured DNA oligonucleotide arrays (Figure 7). These PGA arrays are unique in that the probes are specifically designed at the 5'-end of the open reading frame. To date, most commercial oligonucleotide probe sets have been designed at the 3'-UTR to both facilitate priming with oligo(dT) during reverse transcription, as well as probe design. However the 3'-UTR maintains substantial sequence variability, and methods to clearly predict the 3'-UTR ends are currently under development. Likewise, 5'-UTR ends that lack TATA elements are also heterogeneous and further study is needed to define the boundaries of these elements. Because most users of microarrays are interested in the relative abundances of encoded proteins, we chose the most direct surrogate, protein coding sequences. Not only are protein coding regions the most biologically relevant portion of a mRNA, these regions are clearly definable and avoid the design problems that stem from heterogeneity typically associated with UTR elements. However when designing probes to the protein coding region, care must be taken to ensure a linear representation of labeled cDNA along the mRNA sequence. We have implemented random priming protocols to adequately represent the 5'-open reading frame. Taking this under consideration, OligoPicker has been designed to filter out possible cross-hybridizations stemming from ribosomal RNAs in a total RNA sample. We have implemented this strategy successfully in our laboratory, and have been able to confirm many examples from differential expression data using qPCR demonstrating that a 5'-biased primer design approach is valid.
| ||||||||||||||||||||||||||||||||||