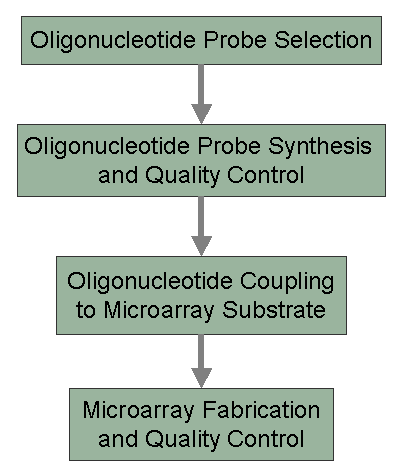
Overview of the Microarray Component of the ParaBioSys PGAThe Microarray Core component of the ParaBioSys PGA has been responsible for the development of oligonucleotide probe sets for DNA microarray production. Because of the high cost associated with commercial probe sets and instrumentation needed for microarray fabrication, the Microarray Core was established to design, synthesize, and produce oligonucleotide probe sets and printed microarrays for the NHLBI academic community. Our goal is to make microarray technology and experimentation more accessible to the inexperienced laboratory by offering cost-effective microarray reagents and services, as well as educational programs in general microarray methodology and data analysis. The following schematic depicts an overview of the processes that the Microarray Core employs for oligonucleotide probe design and synthesis, quality control, and array fabrication. Oligonucleotide probes are designed using the OligoPicker software package developed by the Bioinformatics component of ParaBioSys. Using this versatile software, 70mer oligonucleotide probe sets have been designed for the mouse, human, rat, cow, chicken and pig genomes. The probes are synthesized using standard phospohoramidite methods and are modified at the 5'-terminus to facilitate covalent coupling of a probe to the microarray surface. Each probe is quantified and analyzed by either capillary electrophoresis or mass spectroscopy to ensure that the probe is of the desired length and purity. After quality control analysis, probe sets are made available to the academic community for cost of production. The Microarray Core also produces pre-printed slides that have undergone extensive quality control testing to assess printing performance during production. For a detailed presentation of processes involved in the production of probe sets and microarrays, please select the topic of interest from the scheme below.
Microarray Products of the ParaBioSys PGAPre-printed microarrays currently availableMicroarray Oligonucleotide sets
Public Microarray Data
Bioinformatic ResourcesTutorials in Microarray Data AnalysisMicroarray ServicesMassively Parallel Real Time PCRWe have developed an experimentally validated algorithm for the identification of transcript-specific PCR primers on a genomic scale. Our online database, PrimerBank, has been created for researchers to retrieve primer information for their genes of interest. PrimerBank currently contains 177,516 primers encompassing most known human and mouse genes. The primer design algorithm has been tested by conventional and real-time PCR for a subset of 1,184 primers with a success rate of 99%. We are now building on these preliminary data to generate a comprehensive collection of experimentally validated primers. Such a collection will enhance the abilities of scientists to explore precise changes in expression of their genes of interest, and will allow the initiation of global transcript abundance monitoring by massively parallel PCR methods. |