| Download OligoPicker | Installation Guide | Download Probe Set |
OligoPicker is to help selecting up to five oligo probes for each of the DNA sequences you provided for microarray spotting. You may freely use it in your research with appropriate acknowledgement. OligoPicker is provided "as is" and the author is not responsible for the consequences from using this program. OligoPicker is distributed under GPL license. If you would like to report problems or suggestions, please email Dr. Xiaowei Wang at xwang@molbio.mgh.harvard.edu.
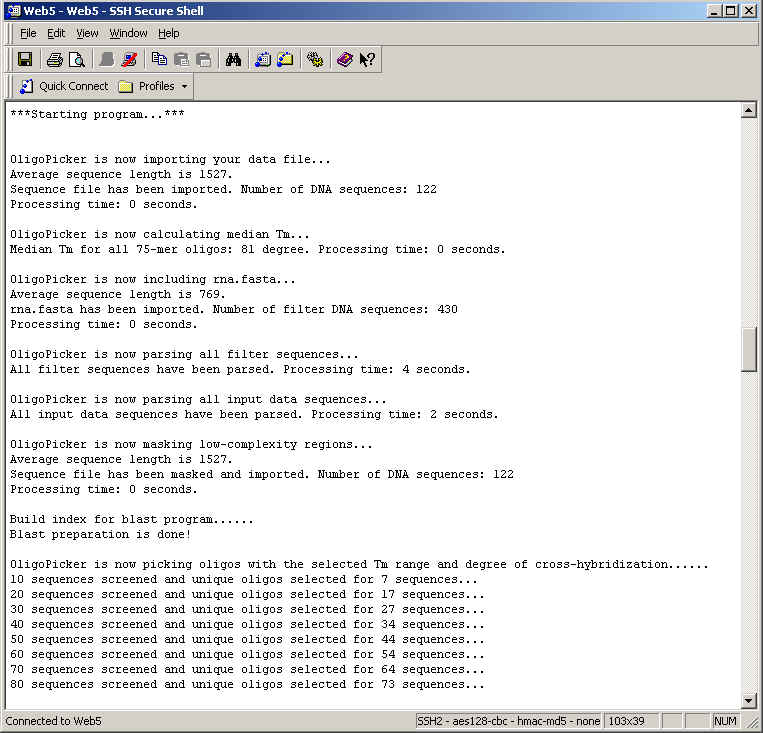
As microarray chips are increasingly used for gene expression studies at genomic scale, it is important to design oligo probes that specifically represent large number of DNA sequences. Cross-hybridization is most likely from the regions with contiguously and perfectly matched bases. Oligo probes should also be sensitive and hybridize under similar conditions. OligoPicker picks specific oligos by skipping regions with contiguous bases common in other sequences. In addition, oligo specificity is double-checked by NCBI BLAST. Sequence regions similar to non-coding RNAs are avoided because total RNA are often used for array hybridization. Low-complexity regions are also filtered out to maintain oligo specificity. Oligos and sequence regions that may form secondary structures are discarded since both the probes and the sequence target sites should be easily accessible for hybridization.
Input data file should be in FASTA format. In a FASTA file, a definition line
is required for each DNA
sequence. Version 2.3.1 or later accepts both NCBI format
(i.e. the
definition line should start with ">gi|...") and non-NCBI FASTA format,
. For example:
>gi|4507798|ref|NM_000462.1| Homo sapiens ubiquitin protein ligase E3A (UBE3A), mRNA
ATGGAGAAGCTGCACCAGTGTTATTGGAAATCAGGAGAACCTCAGTCTGACGACATTGAAGCTAGCCGAA
TGAAGCGAGCAGCTGCAAAGCATCTAATAGAACGCTACTACCACCAGTTAACTGAGGGCTGTGGAAATGA
AGCCTGCACGAATGAGTTTTGTGCTTCCTGTCCAACTTTTCTTCGTATGGATAATAATGCAGCAGCTATT
AAAGCCCTCGAGCTTTATAAGATTAATGCAAAACTCTGTGATCCTCATCCCTCCAAGAAAGGAGCAAGCG
CAGCTTACCTTGAGAACTCGAAAGGTGCCCCCAACAACTCCTGCTCTGAGATAAAAATGAACAAGAAAGG
>gi|3646015|emb|AJ001483.1|HPVADX1L1 human papillomavirus (HPV) related to epidermodysplasia verruciformis HPV (L1 gene) (isolate HPV ADX1)
AACACTAATTTCTGTATCAGTGTCTCCTCAAATGATCAGGCATTACAGGAATACAATACTGCAAACTTTA
GAGAATATTTGAGACATGTAGAAGAGTATGAATTATCCTTTATATTACAATTATGTAAAGTTCCATTAGA
GCCAGAAGTATTAGCACAAATTAATGCTATGAATGCAGACATTTTAGAGGATTGGCAATTAGGTTTTGTT
CCTTCTCCTGACAATCCCATCAATGATACATATAGATACATACATTCAGCAGCCACACGGTGTCCAGATA
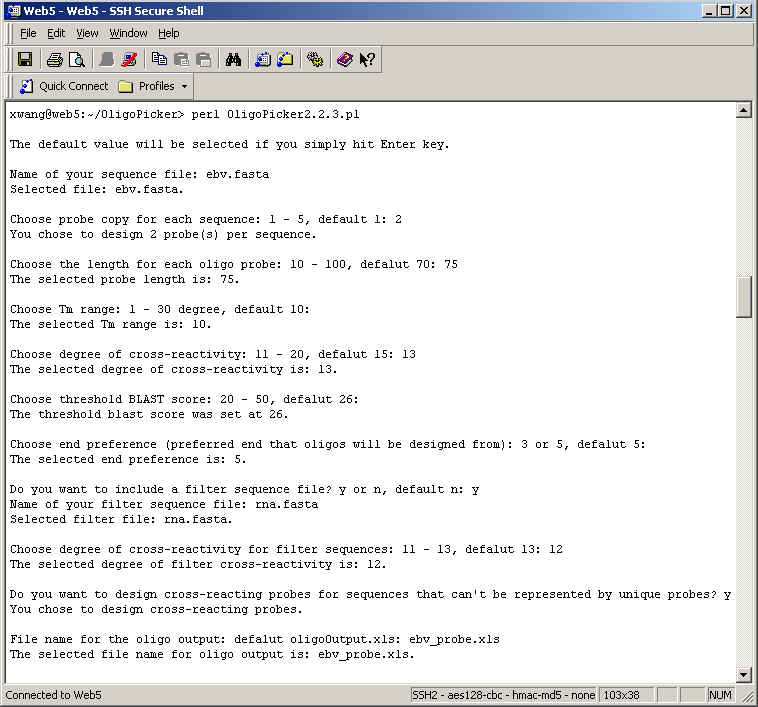
Probe Copy. This specifies how many non-overlapping unique probes are designed for each sequence. Up to five probes may be designed.
Probe Length. This specifies the length for each of the probes in the probe set. The range is 20 - 100 bases and the recommended value is 70.
Tm range. The allowable Tm range is 1 – 30 °C and the default value is set at 10 °C. The screening stringency increases as the Tm range becomes smaller.
Cross-reactivity. This sets the threshold for rejection of contiguous matches. The range is 10 – 20 with the default value set at 15. None of the designed unique oligo probes has any repetitive n-mer in any other input sequence. We believe that stretches of perfectly matched sequences (such as 15-mers) are more likely to cause cross-hybridization than longer homologous sequences with a few mismatches. The default value 15 is preferred because the probability of having two identical 15-mers is one out of 109. This level of stringency should be enough for most sample universe. The smaller the number, the more stringent (the better) condition it is for microarray experiment. However, it is also increasingly difficult to select unique oligos for DNA sequences because of the more stringent selection criteria. As a result, OligoPicker may fail to pick unique oligos for some sequences.
BLAST score. This is supplemental to the n-mer contiguous match screening process. Oligos with NCBI BLAST score higher than the specified value will be discarded. The default value is determined by the threshold size of contiguous matches.
End preference. Probes can be selected from either the 5’ or 3’ end. OligoPicker will start to screen oligos from the end you selected. If the oligos do not qualify, OligoPicker will continue to screen oligos either upstream or downstream from the end until qualified oligos are found.
Filter file (optional). The program allows users to specify a second NCBI FASTA file, e.g. the rna.fasta file provided by OligoPicker, which contains rRNA and snRNA sequences from human, mouse, and rat. All the sequences in this filter file are used during the probe screening process against sequences in the data file. However, oligo probes are not designed for sequences in the filter file. The final oligo probes do not cross-react to any filter sequence.
Cross-reactivity to filter sequences (optional). Oligo cross-reactivity is evaluated against all filter sequences. The concept is the same as cross-reactivity mentioned above to input data sequences. The degree of cross-reactivity to filter sequences can be set here and this value can be different from the degree of cross-reactivity to input data sequences.
Cross-reacting probes (optional). For those sequences that cannot be represented by unique probes, you may optionally design cross-reacting probes for them. For example, there are two splice isoforms A and B. Isoform A is the longer form and is already represented by one unique probe. However, it is impossible to design unique probes for isoform B because the whole sequence of B is part of A. In this case, OligoPicker can design a non-unique probe for B that also cross-reacts to A. By analyzing both the unique probe for A and the non-unique probe for B, you may know the expression levels for these isoforms. Including this option will significantly slow down the speed of OligoPicker.
Output file contains the following information:
A Perl 5 interpreter or higher on a Linux system is required.
| Computer Speed | Number of Sequences | Average Sequence Length | Time Needed | Memory Needed |
| 1.5 GHz | 100 | 1.2 kb | < 1 minute | minimum |
| 1.5 GHz | 20,000 | 1.2 kb | 3 hours | 390 MB |
Click here to download OligoPicker 2.3.2 for Linux platform.

NCBI BLAST and DUST programs were integrated into OligoPicker. We thank NCBI for providing such excellent free tools to the bioinformatics community.
This page was created on 8/13/02 by Xiaowei Wang. Last modified on 5/22/03